January 3, 2019 | Aptamers | Valerie Jones, PhD
Aptamer Selection & Development
What are Aptamers?
Often described as synthetic antibodies, aptamers are single-stranded DNA or RNA molecules that fold into secondary structures which can bind to target molecules with high affinity and specificity. In 1990, the first aptamer was developed against T4 DNA Polymerase using RNA1. Since then, both aptamer selection methods and subsequent applications have expanded significantly. Aptamers can now be developed using both RNA and DNA, as well as non-natural bases, backbones, and small molecules. Applications include: detection molecules in ELISA-like assays, protein-specific tissue staining, targeted drug delivery, and an FDA-approved treatment for macular degeneration2,3.
Aptamers vs. Antibodies: The Aptamer Advantage
With their robust binding affinities and high specificity for small biomolecules, aptamers have generated great interest as an alternative to antibodies. A comparison between the properties of aptamers and antibodies is summarized below.
| APTAMER | ANTIBODY | |
|---|---|---|
| Properties | Higher specificity than antibodies | Higher affinity than aptamers |
| Stability & Storage | Long shelf life at room temperature | Long shelf life when frozen |
| Remains stable after multiple freeze thaws | Degrades after multiple freeze thaws | |
| Can be designed to resist enzymes | Cannot be designed to resist enzymes | |
| Can be "stored" digitally and re-synthesized at low cost | Hybridoma storage and maintenance required | |
| Development & Scalability | 3-5 months (selection & sequencing) | 6-9 months (immunization & hybridoma development) |
| No animals required | Animals required | |
| <1 week to replenish | 3-4 weeks to replenish | |
| Very low batch-to-batch variability | Low to moderate batch-to-batch variability | |
| Target Molecule | Any target may be used | Target limited to immunogenic, non-toxic molecules |
| Poor immunogens or small molecules: no conjugation required to enhance selection | Poor immunogens or small molecules: carrier protein required to enhance immunogenicity | |
| Quantity needed for selection: 100 µg protein / 1 mg small molecule | Quantity needed for immunization: 5 mg protein / 5 mg conjugated peptide | |
| Modification | Easily conjugated to proteins, peptides, drugs, and other small molecules | Easily conjugated to proteins, peptides, drugs, and other small molecules |
| Can be modified with fluorescent dyes | Can be modified with fluorescent dyes | |
| Biotinylation ~$100 / mg | Biotinylation ~$1000 / mg | |
| Cost | Individual aptamer selection: starts at $5,500 | Monoclonal antibody: starts at $4000 |
| "Polyclonal" enriched aptamer pool: $1000 | Polyclonal antibody: starts at $1200 |
SELEX Techniques
Aptamers are developed using a process known as SELEX (Systemic Evolution of Ligands by Exponential Enrichment). During SELEX, trillions of random DNA oligos are mixed with the target molecule, and sequences that bind to the target are then collected and amplified. After multiple rounds of selection, the DNA is sequenced and individual sequences are then evaluated for binding. Using SELEX, aptamers can be developed with affinity for a variety of targets, including proteins, peptides, and small molecules.
In general, SELEX is accomplished in 3 phases.
- Selection: DNA sequences that bind to the target are partitioned from sequences that do not. Binding sequences are amplified via PCR and brought forward for additional rounds of SELEX (generally 5-10).
- Sequencing: RayBiotech uses NGS and data analysis to determine which aptamers to evaluate as opposed to randomly selecting individual clones.
- Candidate Evaluation: Aptamers are evaluated for binding and the best aptamer is supplied to the customer
RayBiotech's custom aptamer service incorporates state-of-the-art techniques into our SELEX process to generate high quality aptamers. Because the selection technique can bias the developing aptamer pool, we will tailor the selection strategy to best suit each customer’s needs. Often this includes using multiple selection techniques within a single project to generate the best aptamers. For example, in many of our selections we will begin with Capillary Electrophoresis SELEX (CE-SELEX) to identify aptamers that bind to all the surfaces of a protein target, enriching the pool of potential binders. This pool of aptamers is then used in a subsequent type of SELEX that more closely resembles the end use assay but does not have as much ability to separate binding from non-binding sequences. Because the pool has already been enriched for binders, we can focus on finding which of those binders work best in your assay.
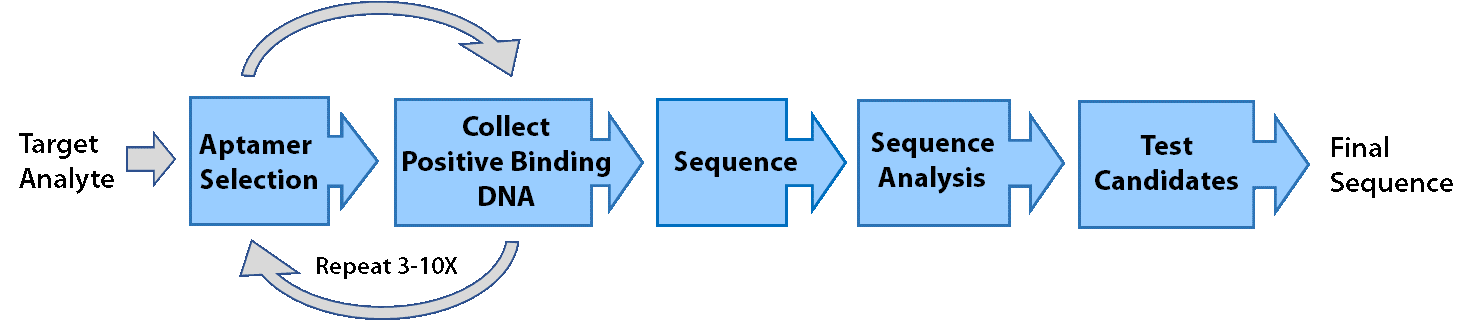
While we can use a wide variety of techniques and variations of selection, they can be categorized into three general types. These techniques are not a comprehensive list of the types of SELEX that have been developed or in use at RayBiotech, and only represent some of the techniques we commonly use:
- Traditional Selection: A traditional selection technique requires the target of interest to be bound to a solid support. This support can be beads, the walls of a microtiter plate, or gel-like resins. The aptamers then bind to the immobilized target and non-binders are removed. The remaining bound aptamers are collected and carried through to the next round. This type of selection is best suited to larger targets such as peptides and proteins, although other targets can be selected for as well. The benefit of this selection technique is that it offers good separation between binding and non-binding DNA sequences, but at the cost of potentially identifying aptamers against the linker or the support that the target is bound to. Additionally, with smaller targets, this technique may limit the number of surfaces available for aptamer binding.
- Reverse Selection: This is similar to traditional selection except that the aptamer library is bound to the solid support, which is then challenged with the target of interest. Depending on the selection strategy, the bound aptamers can be collected by secondary capture of the target or by releasing the bound aptamers from their solid support. A benefit of reverse selection is that the target of interest is not bound or modified in any way, allowing the aptamer to recognize the target in its native form, making this type of selection ideal for smaller molecules. However, efficiency of separation between binding and non-binding DNA sequences is generally lower than other types of selection, resulting in longer selection times.
- CE Selection: The traditional method requires the target to be bound to a solid support, which potentially alters the target the aptamer is selected against. However, at RayBiotech we use CE to separate binding sequences, permitting the DNA to interact with the free protein in solution. This allows the two molecules to recapitulate natural interactions in serum or other fluids. Additionally, the separation efficiency of CE selection is very high which means fewer rounds of selection are required to enrich the aptamer pool. While traditional SELEX can require 10 rounds of selection or more, CE can effectively identify aptamers in as few as 3 rounds, which helps to reduce the effects of PCR bias on the selection4,5. The use of CE also allows us to monitor the development of the aptamer library throughout the selection process.
In addition to a variety of selection techniques, RayBiotech uses Next Generation Sequencing (NGS) instead of plasmid cloning techniques to identify individual aptamers. We utilize the data gained in NGS to analyze the entire aptamer pool and identify common aptamer sequences, motifs, and other patterns in the library. Using this information allows us to use data analytics to identify better aptamer candidates than those identified by randomly selected clones, increasing the likelihood of success.
After candidate sequences are identified, they are synthesized and assessed for their ability to bind the target of interest. The techniques used may vary based on the target and the final application of the aptamer. As with antibodies, the application can impact the aptamer’s performance and thus multiple techniques maybe used to determine the functionality of the final aptamer.
- Tuerk, Craig, and Larry Gold. "Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase." Science 249.4968 (1990): 505-510.
- Vinores, Stanley A. "Pegaptanib in the treatment of wet, age-related macular degeneration." International journal of nanomedicine 1.3 (2006): 263.
- Bauer, Michelle, et al. "The Application of aptamers for immunohistochemistry." Nucleic acid therapeutics 26.3 (2016): 120-126.
- Stuart, Christopher H., et al. "Selection of a Novel Aptamer Against Vitronectin Using Capillary Electrophoresis and Next Generation Sequencing." Molecular Therapy—Nucleic Acids 5.11 (2016): e386.
- Mosing, Renee K., and Michael T. Bowser. "Isolating aptamers using capillary electrophoresis–SELEX (CE–SELEX)." Nucleic Acid and Peptide Aptamers: Methods and Protocols (2009): 33-43.
13 Feedbacks on "Aptamer Selection & Development"